文章信息
文章题目:Genomic atlas of 8,105 accessions reveals stepwise domestication, global dissemination, and improvement trajectories in soybean
期刊:Cell
发表时间:2025 年 10 月 1 日
主要内容:崖州湾国家实验室田志喜团队通过对 8105 份大豆种质进行全基因组分析,首次明确黑大豆是大豆驯化的关键中间类型,并揭示其逐步驯化路径与两大起源中心(黄淮地区与西北地区)。研究系统解析了大豆在全球传播过程中的适应性基因选择与中国育种目标的时代变迁,构建了首个大豆 QTN 库与在线变异数据库,为未来高产、高油、高蛋白大豆的精准育种提供了重要基因资源与平台。
原文链接:
https://www.cell.com/cell/fulltext/S0092-8674(25)01038-4
使用TransGen产品:
pEASY®-Uni Seamless Cloning and Assembly Kit (CU101)
ProteinFind® Anti-GFP Mouse Monoclonal Antibody (HT801)

研究背景
现代栽培大豆约在 5000-6000 年前由野生大豆在中国被驯化,随后逐步传播至亚洲、欧洲、北美、南美等不同国家和地区,为人类和动物提供了主要的植物油脂和蛋白质来源,已经成为全球最重要的粮油作物之一。作为重要的豆科作物,大豆在长期驯化、传播和改良过程中,其种质资源在世界各地逐渐形成了丰富的多样性,在植株形态、生态适应性、生产特性等方面都发生了巨大变化和分化。然而,关于大豆种质资源演化的一些重要问题并不清楚,解析大豆种质资源遗传基础,深刻理解性状形成的决定基因,对大豆种质资源利用和优良品种培育具有重要意义。
文章概述
研究团队全面调查了 8105 份大豆材料的进化轨迹,包括野生近缘种、地方品种和改良品种,以阐明大豆驯化、传播和改良的过程。研究发现黑大豆是大豆驯化历史上的一个重要中间体,并揭示了性状和基因的逐步选择。还阐明了所选基因在大豆传播和改良中的等位基因多样性。此外,构建了一个包含 8105 份大豆材料的变异图谱,并建立了一个大豆数量性状核苷酸 (QTN) 文库和一个选定基因的在线遗传变异数据库。本研究绘制了一幅清晰的大豆在驯化、传播、改良过程中的关键基因选择的地理和历史性“画卷”图谱,并提出了新的大豆起源假说。相关发现不仅为理解作物驯化与改良的一般规律提供了新的视角,也为充分利用种质资源开展分子设计育种奠定了坚实的理论基础和数据平台。
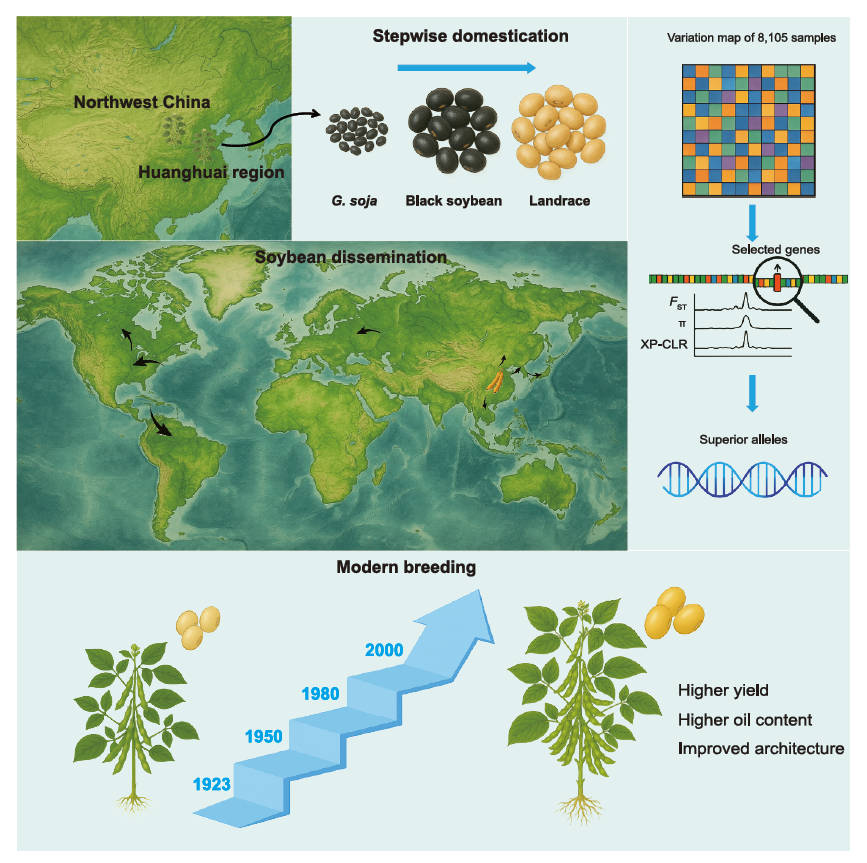
大豆的驯化、传播与改良过程
全式金生物产品支撑
优质的试剂是科学研究的利器。全式金生物的通用版同源重组无缝克隆试剂盒(CU101)、抗 GFP 标签鼠克隆抗体(HT801)助力本研究。产品自上市以来,凭借优异的性能,深受客户青睐,多次荣登知名期刊,助力科学研究。
pEASY®-Uni Seamless Cloning and Assembly Kit(CU101)
本产品利用特殊的重组酶和同源重组的原理,可以将任意方法线性化后的载体和与其两端具有 15-25 bp 重叠区域的 PCR 片段定向重组,可以实现最多 15 个片段的高效无缝拼接。
产品特点
• 阳性率高,克隆数多。
• 高效连接:最高可实现 15 个片段无缝连接。
• 快速重组:5-15 min 即可完成反应。
• 大容量组装:可成功构建 31.8 kb 的质粒 (载体+片段)。
• 长载体组装:支持 14 kb 长载体组装。
• 广谱兼容:支持低浓度 (0.003 pmol) 单片段、多片段高效连接。
ProteinFind® Anti-GFP Mouse Monoclonal Antibody(HT801)
抗 GFP 标签鼠单克隆抗体为高纯度的抗小鼠单克隆抗体,属 IgG1 同型,免疫原为人工合成的全长 GFP 蛋白。
产品特点
• 高纯度的抗小鼠单克隆抗体,特异性强。
• 高度特异识别重组蛋白 C 末端或 N 末端的 GFP 标签。
• 适用于定性或定量检测 GFP 融合表达蛋白。
使用 pEASY®-Uni Seamless Cloning and Assembly Kit(CU101)产品发表的部分文章:
• Xu Y, Zhu T F. Mirror-image T7 transcription of chirally inverted ribosomal and functional RNAs[J]. Science, 2022.(IF 63.71)
• Zhu Z, Wang Y, Liu S, et al. Genomic atlas of 8,105 accessions reveals stepwise domestication, global dissemination, and improvement trajectories in soybean[J]. Cell, 2025.(IF 42.50)
• Shi J, Mei C, Ge F, et al. Resistance to Striga parasitism through reduction of strigolactone exudation[J]. Cell, 2025.(IF 42.50)
• Bai X, Sun P, Wang X, et al. Structure and dynamics of the EGFR/HER2 heterodimer[J]. Cell Discovery, 2023.(IF 38.07)
• Wang H, Yang J, Cai Y, et al. Macrophages suppress cardiac reprogramming of fibroblasts in vivo via IFN-mediated intercellular self-stimulating circuit[J]. Protein & Cell, 2024.(IF 21.10)
• Xu J, Liang Y, Li N, et al. Clathrin-associated carriers enable recycling through a kiss-and-run mechanism[J]. Nature Cell Biology, 2024.(IF 17.30)
• Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity[J]. Nature communications, 2021.(IF 14.92)
• Shi C, Yang X, Hou Y, et al. USP15 promotes cGAS activation through deubiquitylation and liquid condensation[J]. Nucleic Acids Research, 2022.(IF 14.90)
• Wang J, An Z, Wu Z, et al. Spatial organization of PI3K-PI (3, 4, 5) P3-AKT signaling by focal adhesions[J]. Molecular Cell, 2024.(IF 14.50)
• Liu S, Fan L, Liu Z, et al. A Pd1–Ps–P1 feedback loop controls pubescence density in soybean[J]. Molecular plant, 2020.(IF 12.08)
• Jin Q, Yang X, Gou S, et al. Double knock-in pig models with elements of binary Tet-On and phiC31 integrase systems for controllable and switchable gene expression[J]. Science China Life Sciences, 2022.(IF 10.37)
• Mu S, Chen H, Li Q, et al. Enhancing prime editor flexibility with coiled-coil heterodimers[J]. Genome Biology, 2024.(IF 10.10)
• Du G, Xiong L, Li X, et al. Peroxisome elevation induces stem cell differentiation and intestinal epithelial repair[J]. Developmental cell, 2020. (IF 10.09)
• Tang Y, Gao C C, Gao Y, et al. OsNSUN2-mediated 5-methylcytosine mRNA modification enhances rice adaptation to high temperature[J]. Developmental cell, 2020.(IF 10.09)
使用 ProteinFind® Anti-GFP Mouse Monoclonal Antibody(HT801)产品发表的部分文章:
• Wu M, Bian X, Huang B, et al. HD-Zip proteins modify floral structures for self-pollination in tomato[J]. Science, 2024.(IF 56.90)
• Zhu Z, Wang Y, Liu S, et al. Genomic atlas of 8,105 accessions reveals stepwise domestication, global dissemination, and improvement trajectories in soybean[J]. Cell, 2025.(IF 42.50)
• Zeng R, Shi Y, Guo L, et al. A natural variant of COOL1 gene enhances cold tolerance for high-latitude adaptation in maize[J]. Cell, 2025.(IF 42.50)
• Ma X J, Wang W, Zhang J Y, et al. NRT1.1B acts as an abscisic acid receptor in integrating compound environmental cues for plants[J]. Cell, 2025.(IF 42.50)
• Li Y, Zhang Z, Chen J, et al. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1[J]. Nature, 2018.(IF 41.00)
• Zhao S, Makarova K S, Zheng W, et al. Widespread photosynthesis reaction centre barrel proteins are necessary for haloarchaeal cell division[J]. Nature Microbiology, 2024.(IF 28.30)
• Fan H, Quan S, Ye Q, et al. A molecular framework underlying low-nitrogen-induced early leaf senescence in Arabidopsis thaliana[J]. Molecular Plant, 2023.(IF 27.50)
• Shi Q, Xia Y, Wang Q, et al. Phytochrome B interacts with LIGULELESS1 to control plant architecture and density tolerance in maize[J]. Molecular plant, 2024.(IF 17.10)
• Wang J D, Wang J, Huang L C, et al. ABA-mediated regulation of rice grain quality and seed dormancy via the NF-YB1-SLRL2-bHLH144 Module[J]. Nature Communications, 2024.(IF 14.70)
• Jia X, Lin L, Guo S, et al. CLASP-mediated competitive binding in protein condensates directs microtubule growth[J]. Nature Communications, 2024.(IF 14.70)
• Chang J, Wu S, You T, et al. Spatiotemporal formation of glands in plants is modulated by MYB-like transcription factors[J]. Nature Communications, 2024.(IF 14.70)
• Zhang H, Huang C, Gao C, et al. Evolutionary-Distinct Viral Proteins Subvert Rice Broad-Spectrum Antiviral Immunity Mediated by the RAV15-MYC2 Module[J]. Advanced Science, 2025.(IF 14.30)
• Du D, Li Z, Jiang Z, et al. The Transcription Factor WFZP Interacts with the Chromatin Remodeler TaSYD to Regulate Root Architecture and Nitrogen Uptake Efficiency in Wheat[J]. Advanced Science, 2025.(IF 14.10)
• Meng T, Chen X, He Z, et al. ATP9A deficiency causes ADHD and aberrant endosomal recycling via modulating RAB5 and RAB11 activity[J]. Molecular Psychiatry, 2023.(IF 13.43)
• Li Y, Du Y, Huai J, et al. The RNA helicase UAP56 and the E3 ubiquitin ligase COP1 coordinately regulate alternative splicing to repress photomorphogenesis in Arabidopsis[J]. The Plant Cell, 2022.(IF 12.00)
• Shen S Y, Ma M, Bai C, et al. Optimizing rice grain size by attenuating phosphorylation-triggered functional impairment of a chromatin modifier ternary complex[J]. Developmental Cell, 2024.(IF 11.80)
• Du D, Li Z, Yuan J, et al. The TaWAK2-TaNAL1-TaDST pathway regulates leaf width via cytokinin signaling in wheat[J]. Science Advances, 2024.(IF 11.70)







